- This topic has 12 replies, 7 voices, and was last updated 3 years, 5 months ago by
 Wirichada Pan-ngum.
Wirichada Pan-ngum.
-
AuthorPosts
-
-
2022-08-20 at 11:11 am #37741
 Wirichada Pan-ngumKeymaster
Wirichada Pan-ngumKeymasterBy the end of the week, I would like you to submit to the discussion board.
-What model structure would the disease you are interested be and please start adjusting the R code to that structure.
– Please state the key characteristics of the disease you are focusing on e.g. transmission, pathogenicity, symptoms, control measures etc. You can write it as text or start using the parameter table in the template provided. Anything you want to let me know about the disease you are going to work on would be useful. (10 points)
Variable name description Value or range Source of information/data aa bb -
2022-09-11 at 4:45 am #38023
Wichayapat Thongrattana
ParticipantFor my study on Tomato Flu, I plan to use the SIR model.
There is a change on scope of work to focus on only Kerala, a state in India, since it is a solely state where Tomato flu case reports are publicly available.
According to (https://www.bmj.com/content/378/bmj.o2101.long), Tomato flu, a self-limited disease, is caused by enterovirus(EV) Coxsackievirus A16(CA16). As it is confirmed as the variant of Hand, Foot, and Mouth Disease(HFMD), it has same mode of transmission spreading through faces and fluid in the spots. Moreover, clinician in Kerala had confirmed that it is not transmitted by mosquitoes.Differently from HFMD, Tomato Flu show sign of symptoms as presence of tomato-like red welts all over the body. The control measure for this disease is follow same procedure as in HFMD which isolated the patient for 2 weeks since the disease will resolve without treatment within 1-2 weeks after infections and there is no to record a single death on this disease.
These are the variables which I have discovered from my research on Tomato Flu.
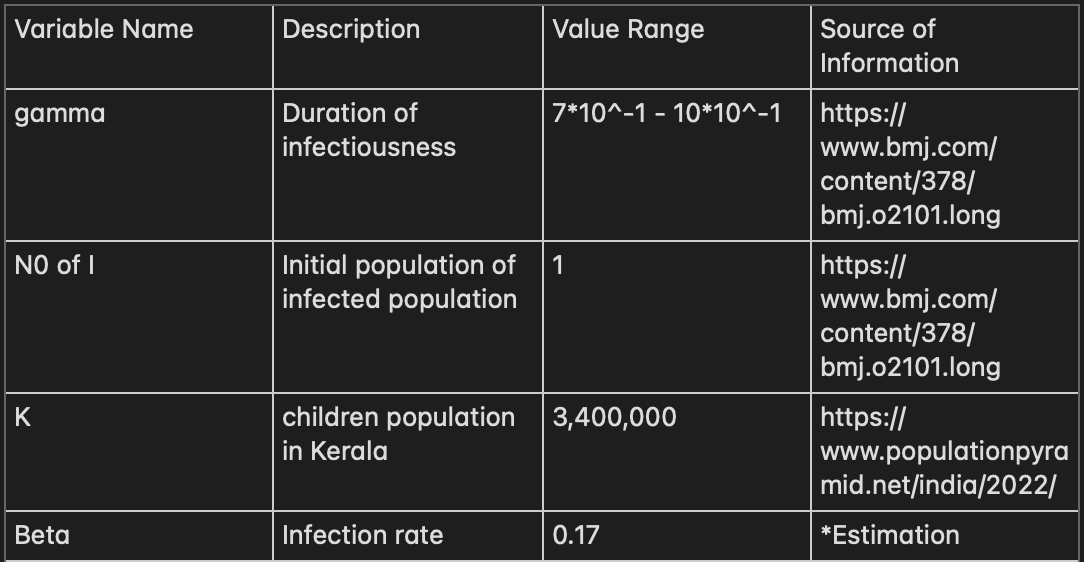
*Beta is estimated based on the compartmental model along with the case report history of Tomato flu as reported of 82 cases after 82 days from first reported case. I have fitted all of the parameter to SIR model and rearrange the value of Beta until N(82) of Infectious match to the actual case report where the number of cases is beginning to rise. From this, I have calculated reproduction number from estimated Beta and averaged Gamma resulting in ~1.5 R0 which closely to reproductive number of HFMD in China between 2008 and 2009 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6805791/).
-
2022-09-15 at 5:50 am #38061
 Wirichada Pan-ngumKeymaster
Wirichada Pan-ngumKeymasterI think gamma is probably the recovery rate (per day) and should actually be 1/duration of infectiousness. OK, it would be nice to now construct and run the model in R.
-
-
2022-09-11 at 5:31 pm #38026
 Arwin Jerome Manalo OndaParticipant
Arwin Jerome Manalo OndaParticipantIn the paper of David & Gomez (2014), they used the SI model for HIV. I think it is rational to use the SI model since the states of HIV can be susceptible and infected. Although there are accounts of about patients recovering from HIV, these cases are very rare – in fact, these remain a huge mystery for scientists. In addition, although there are HIV medications that can suppress the viral load to undetectable levels and proclaim one to be “HIV-undetectable” who cannot transmit the disease, incorporating a “recovered” state would be erroneous as the patient is not completely cured from the disease.
Variables:
1. Beta-1, Beta-2, Beta-3: Transmission rates for each gender (heterosexual, bisexual, homosexual), respectively. Data: Infections rates estimated by David & Gomez (2014) (from 1990-2012) and I am continuing search for updated values or if there are published values in literature.2. Infected (I): Initial number of infected people
Data: 56982 people with HIV registered taking anti-retroviral therapy
Source: https://doh.gov.ph/sites/default/files/statistics/EB_HARP_January_AIDSreg2022.pdf3. Susceptible (S)***: Number of susceptible people
Data: 72.02 million
Sources:
https://data.worldbank.org/indicator/SP.POP.1564.TO.ZS?locations=PH
https://popcom.gov.ph/popcom-number-of-filipinos-in-2021-estimated-at-110-8-million-sizes-of-families-trending-lower-at-4-members/#:~:text=POPCOM%3A%20Number%20of%20Filipinos%20in,Commission%20on%20Population%20and%20Development
https://doh.gov.ph/sites/default/files/statistics/EB_HARP_January_AIDSreg2022.pdf***Of the 110.8 million Filipino people (source: Philippine POPCOM), 65% are in the age of 15-64 years old (source: WorldBank). It is assumed that this population group are comprised of people actively having sex. David & Gomez (2014) also looked at this specific population group.
-
2022-09-11 at 7:11 pm #38027
 Karina Dian LestariParticipant
Karina Dian LestariParticipantFor modelling the risk of malaria for travelers to plantation area inside the forest in Rejang Lebong district, I will use the same compartments described in the following paper (https://malariajournal.biomedcentral.com/articles/10.1186/1475-2875-8-296#MOESM1). The only difference I made is I do not incorporate Latent Mosquito compartment, therefore the compartments I will use are:
With probe represent a cohort of travelers. The model’s parameters are:
The value of model parameters will be searched from several literatures that are relevant to malaria in Indonesia situation.
-
2022-09-15 at 6:03 am #38062
 Wirichada Pan-ngumKeymaster
Wirichada Pan-ngumKeymasterKarina, it would be good to mention more about the setting. I am not clear if we focus on falciparum here? What do we assume about the immunity among the travelers? I am curious to see the model adjusted to your questions and setting next.
-
2022-09-20 at 3:40 pm #38192
 Karina Dian LestariParticipant
Karina Dian LestariParticipantYes, we will be focusing on Pf infection and the travellers are assumed to be naive to malaria in the model.
-
-
-
2022-09-15 at 6:06 am #38063
 Wirichada Pan-ngumKeymaster
Wirichada Pan-ngumKeymasterArwin, somehow I cannot click reply next to your post. I think you have captured the important elements of HIV there and also aware of what assumptions you made to have a simple version of the model. For the final project, you can spell out the parameter estimates into table with the references.
-
2022-09-19 at 10:42 am #38160
 Arwin Jerome Manalo OndaParticipant
Arwin Jerome Manalo OndaParticipantThank you Professor.
-
-
2022-09-15 at 11:32 pm #38091
 Napisa Freya SawamiphakParticipant
Napisa Freya SawamiphakParticipantI will use SIR model for the mathematical modeling of HBV transmission dynamic in Thailand regardless of vaccination and transmission route. The variables include S = susceptible, I = HBV infected, and R = recovered people (as suggested by https://science.buu.ac.th/ojs246/index.php/sci/article/view/1623/1551 ). Also, I is subclassified between acute infection and chronic infection; IA= early HBV infected and IC= Chronic/late HBV infected. As HBV infection is permanent, recovered individuals will define as having non-symptoms related to HBV (controlled symptoms).
I can not attach the table and model . Please access the full report here https://tinyurl.com/3yjcs4bn
Thank you.
-
2022-09-17 at 6:19 am #38132
 Wirichada Pan-ngumKeymaster
Wirichada Pan-ngumKeymasterOk, great start. Now HBV main transmission is vertical route. Can you think how this may be added into the model since we want to control it?
-
-
2022-09-20 at 12:53 am #38168
 Ashaya.iParticipant
Ashaya.iParticipantI would like to use SIR model to the study the dynamics of COVID-19 and bacterial pneumonia coinfection in Thailand. S. Ansari et al. stated that bacterial or fungal coinfections are unlikely to be common in patients with mild COVID-19 when compared with those with more severe disease upon admission to the hospital. According to R. Kumar et al, the study of coinfections in a pandemic situation such as COVID-19 has become imperative due to the clinical, diagnostic and therapeutic challenges. To reduce the risk of co-infection along with a COVID-19 infection, such as influenza and pneumococcus, adults and children—especially those with underlying health conditions—the elderly, and those with weakened immune systems should receive all vaccinations as scheduled. Therefore, study of the dynamics of COVID-19 and bacterial pneumonia coinfection is necessary to the effective patient management and treatment plan.
The parameters of this model can be included;
Λ: recruitment rate of susceptible population.
µ: natural death rate.
α: transmission rate of COVID-19.
γ: recovery rate of people infected with COVID-19 and bacterial pneumonia coinfection
δ: death rate of COVID-19 and bacterial pneumonia coinfection-induced
σ: rate of loss of immunity against COVID-19 infection
θ: rate of loss of immunity against pneumococcal pneumonia
Source: https://arxiv.org/abs/2207.13265-
2022-10-01 at 9:20 pm #38488
 Wirichada Pan-ngumKeymaster
Wirichada Pan-ngumKeymasterOk. Look forward to reading the full report and will give some detailed feedback. Thanks!
-
-
-
AuthorPosts
You must be logged in to reply to this topic. Login here


