Forum Replies Created
-
AuthorPosts
-
-
2025-11-05 at 2:24 pm #51807
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHello Nattha,
We are so sorry for the misleading title. We used to have a final project presentation when the course format was in-person. There will be no presentation required for our class.
As for your other question. Results that did not turn out well are also acceptable for the final project.
Best of luck,
Pimwadee -
2025-10-28 at 11:07 am #51610
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterEven though we would love to see you complete data preparation and basic statistics in R or Orange, we realize that students may have preferences and/or fluency in other software packages. Since the data preparation and basic statistics are not the main focus of this course, you are allowed to use Power Query and SPSS for those steps. The main data mining algorithms, however, must be completed with the software R or Orange. I hope this helps!
Pimwadee
-
2025-10-17 at 12:41 pm #51435
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterYes, Kaggle is a perfectly acceptable source for project data.
Aj.Pimwadee
-
2025-10-02 at 1:38 pm #51110
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHello,
The cross-validation is especially beneficial for small datasets because it allows a more robust model evaluation when data size is small. On the other hand, large datasets can be split into just training and testing and be trained and tested accordingly. That satisfies the principle of testing performance of a model using unseen data (i.e. test data). However, that’s not to say that cross-validation cannot be used for large datasets. Many data practitioners evaluate models on cross-validation when they can despite their sizeable datasets. Cross-validation is a technique to split data for training and testing various times, and therefore is useful in model evalution.
I hope this helps.
-Pimwadee
-
2025-09-23 at 1:19 pm #50785
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterWhat a great question. Learning about multiple algorithms all at once can be overwhelming. Let’s try to tackle this.
When applying clustering to real-world data, we need to ask ourselves this question: how much do we already know about our data? Do we expect to see hierarchies in data? For example, we might be studying rocks and want to group them by their properties. There can be smaller groups of rocks with specific properties, which can then form broader groups of rocks with more common properties. In this case, we might consider AGNES as our algorithm. Then, there’s also a general-purpose algorithm like k-means. However, if you suspect that your data has specific shapes, it will not work well with partitioning algorithms like k-means or k-variant algorithms (which will try to capture data in spherical clusters), and you might consider density-based algorithms instead. To know that data is expected to have specific shapes requires some form of background knowledge.
All of that said, we should also consider pros and cons for each technique when choosing clustering algorithms. Partitioning methods require a k parameter to run, while hierarchical methods can take a long time to run. Many advanced algorithms can handle specific needs, such as handling overlapping clusters. Many algorithms take a long time to run for large datasets due to their complexities. With background information and/or a hunch about the data, coupled with our knowledge for each type of algorithm, we are equipped to make a decision on which algorithms to use.
-
2024-10-06 at 12:19 am #45787
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHi,
The data link might have been changed. Please download the data from the website listed in task #1 instead. Follow the link, then click download button located on the top right of the screen.
I hope this helps.
Pimwadee
-
2023-10-22 at 11:33 am #42506
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHi Siriphak,
Thank you for your question. I would use unique() to list all possible answers for the data.
You may also use str() or summary() on your data.
I hope that helps.
Pimwadee
-
2023-10-16 at 6:55 am #42379
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHi Tanyawat,
I’m sorry it took a while for me to circle back to answering this question in written form.
1) An answer for this question is highly data-dependent. But the rules of thumb for data training is the more the better. And then you set that aside for training 70-80% of all the data you have.
If there are clear patterns hidden in your data, then not much data is needed for training (as clearly seen in our sample dataset from the lectures). If you find that even the large amount of data could not produce satisfying decision trees, perhaps the right attributes for prediction are not present in your data, or perhaps they were there but there were confounding effects at play, in which case a preprocessing of data might lead to better results.
This idea applies to the appropriate number of attributes. If you have the “right” attributes, then you will not need many of them to predict the data. But, of course, real-life data usually are not perfect. Another thing to consider is the more number of attributes are being fed into the algorithms, the more time it takes and the more complex the resulting decision trees will be.
2) The decision tree algorithm that we use in class for r (rpart) should be able to handle this. For other tools / programming languages / software, you will need to read the documentation of that specific tool/language/software to see if they have this ability embedded.
Regards,
Pimwadee -
2023-10-08 at 4:26 am #42199
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHi Abdillah,
You are right to be concerned about outliers. Outliers are ubiquitous in all kinds of datasets! When we are trying to make sense of something, we probably don’t want to think too much about exceptional cases like outliers. Because outliers can distort our understanding of the nature of the data.
There is an exception if we want to study those extreme cases, then outliers are important. But for clustering, algorithms that are prone to outliers like k-means or hierarchical clustering need to be treated with care. If we are aware that data has outliers, some may choose to play with inclusion / exclusion of those cases to see if clustering results change.
I hope that makes sense. Thanks for a spark of discussion!
Pimwadee
-
2020-09-08 at 9:38 pm #22316
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterThank you for informing us. I have chosen the README.md link. I replaced it with the dead link on the assignment page.
-
2020-08-27 at 2:47 pm #22037
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterThe dataset I believe has been processed from its original. The data with a bunch of numbers with no B or M diagnosis is in fact described here. http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/breast-cancer-wisconsin.names
An excerpt from the above file is below:
=========================
7. Attribute Information: (class attribute has been moved to last column)# Attribute Domain
— —————————————–
1. Sample code number id number
2. Clump Thickness 1 – 10
3. Uniformity of Cell Size 1 – 10
4. Uniformity of Cell Shape 1 – 10
5. Marginal Adhesion 1 – 10
6. Single Epithelial Cell Size 1 – 10
7. Bare Nuclei 1 – 10
8. Bland Chromatin 1 – 10
9. Normal Nucleoli 1 – 10
10. Mitoses 1 – 10
11. Class: (2 for benign, 4 for malignant) -
2020-08-23 at 12:17 pm #21968
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterReferring to the file “breast-cancer-wisconsin.names” within this data folder (http://archive.ics.uci.edu/ml/machine-learning-databases/breast-cancer-wisconsin/), the class attribute is in the last column.
-
2020-08-19 at 8:51 pm #21832
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHi. It seems from the error message (NA/NaN/Inf in foreign function call (arg 1)) that the error has something to do with invalid data types. Perhaps the input data is not conforming to the required input for kmeans?
-
2020-08-19 at 3:04 pm #21821
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterThanks for the question! Yes, the reading assignment 2 of section 1.5 mentions both single linkage and complete linkage, which are two different ways for an agglomerative clustering to merge smaller clusters to bigger clusters. Even though we did not mean to elaborate on their definitions and difference in this course, we are happy to see its discussion on this forum.
To recap, single linkage clustering will merge based on the smallest distance of members within clusters, while complete linkage clustering will do so based on the largest distance of the members within clusters. That yields very different results as you have noticed.
By its definition, the single linkage will give you the merge which spreads locally. It will merge anything (points or clusters) that comes as the next closest objects together. I’m thinking it’s similar to water droplets being merged on a surface. Once smaller droplets form bigger water droplets, they expand to the next closest water droplet to merge. Edge-to-edge distances between clusters are a deciding factor for merging here.
In contrast, the complete linkage looks for a minimum farthest distance between clusters. This makes it not sensitive to outliers, i.e. it will not merge by only looking locally for the closest neighbor, but it will look more globally for who is the most similar neighbor. As a result, the complete linkage tends to give us more compact clusters. That is not to say that the single linkage is without its merit. The single linkage can produce clusters that are more inclusive of special cases.
And you can decide the type of clustering you want for your analysis depending on your goal of analysis as well as the nature of data.
-
2021-10-30 at 2:57 pm #32656
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterFor the purpose of this course, you do not need to compare your tree model with these methods. Since our main objective here is to learn and practice decision tree, it is all you need. We do not require you to know other methods not covered in the scope of this course.
-
2021-10-18 at 10:32 am #32236
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterHi Navinee,
That is a good question. The short answer is there is no agreeable number in the industry as to which between_SS / total_SS is considered high or low. But let me take you through some examples, which I found on this link. It is helpful to consider between_SS / total_SS altogether with some plots of the data.
Now please consider case 1. This clustering yields between_SS / total_SS to be 47.5%. And the plot is displayed below.
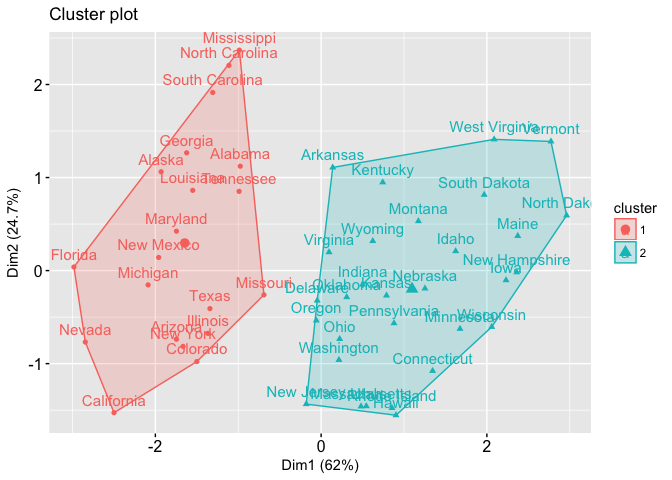
Whereas in case 2, you get between_SS / total_SS to be 71.2 %. Below is the plot of the second case.
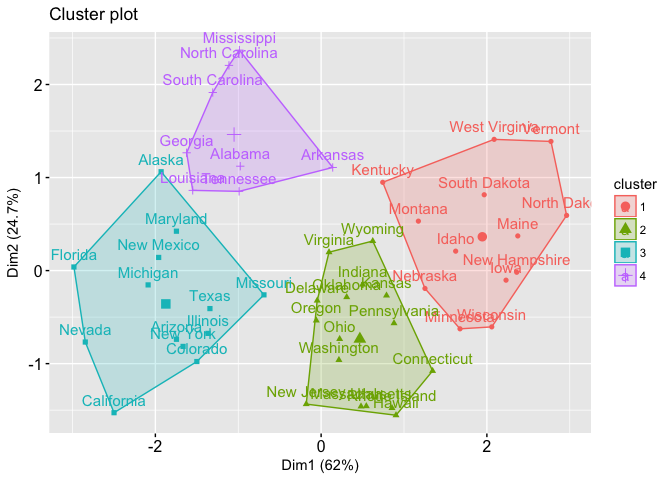
In the first case, 47.5% shows a good enough result, the clusters are somewhat separated. But I am not sure if we take the colors away from the plot we can still clearly see the clusters. Whereas in the second case, 71.2% shows more well-separated clusters (there’s more white space between clusters).
So in my opinion, if the between_SS / total_SS is over 70% I usually considered this very good, which means if I were to plot the data I will see the clusters nicely split. If my between_SS / total_SS is between 50-70%, I still consider this quite OK. I might want to verify the cluster qualities with other numbers and maybe with plots of data. But sometimes that’s as much as you can get out of your data. If my between_SS / total_SS is below 30%, then I think the result is not very good.
I hope this helps. If you have more questions, please let us know.
-
2021-10-07 at 7:58 pm #31955
 Pimwadee ChaovalitKeymaster
Pimwadee ChaovalitKeymasterDear Rawinan,
Thanks for your question. Accuracy is the percentage of correct prediction, while error rate is the percentage of the incorrect prediction. Those two numbers are calculated across all classes. Precision tells you how correct the algorithm was in predicting the positive class. Finally, recall tells you how much of the actual positive cases that the algorithm was able to predict as positive. I also did answer in Thai during the session so you may check out the video recording of the class meeting.
Pimwadee
-
-
AuthorPosts
